I am currently a postdoctoral associate in the Department of Biostatistics at Yale University, advised by Prof. Hongyu Zhao. I completed my Ph.D. at the Hong Kong University of Science and Technology in 2023, under the supervision of Prof. Can Yang and Prof. Yang Wang.
My research interests focus on the development of machine learning and AI methodologies, with applications in computational biology and biomedical data science.
Selected Publications
* Joint first authors. # Corresponding authors. † Consortium membership.
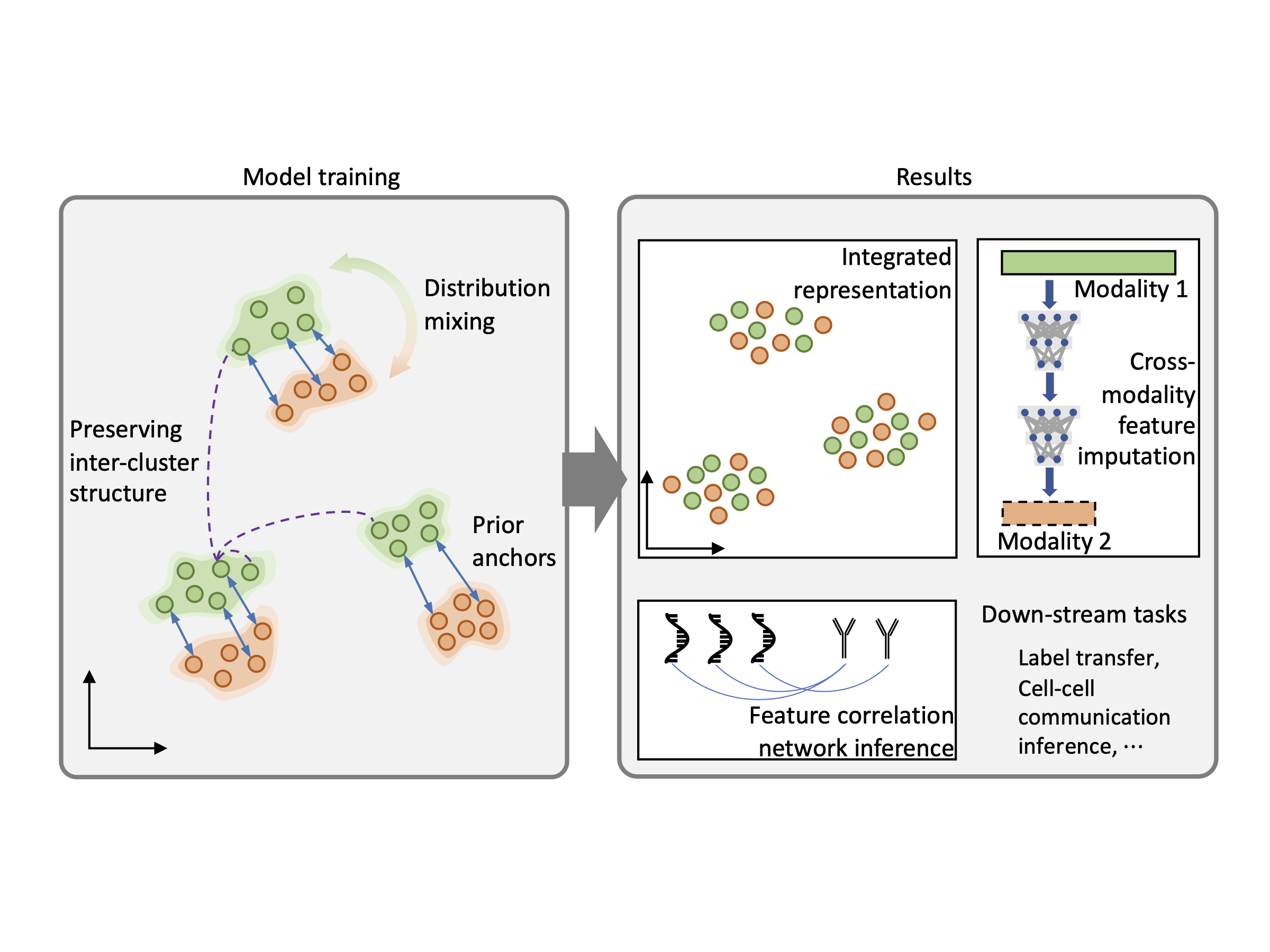
scMODAL: A general deep learning framework for comprehensive single-cell multi-omics data alignment with feature links Gefei Wang, Jia Zhao, Yingxin Lin, Tianyu Liu, Yize Zhao, Hongyu Zhao#.
Nature Communications. 2025.
Paper |
bioRxiv |
Software |
Website 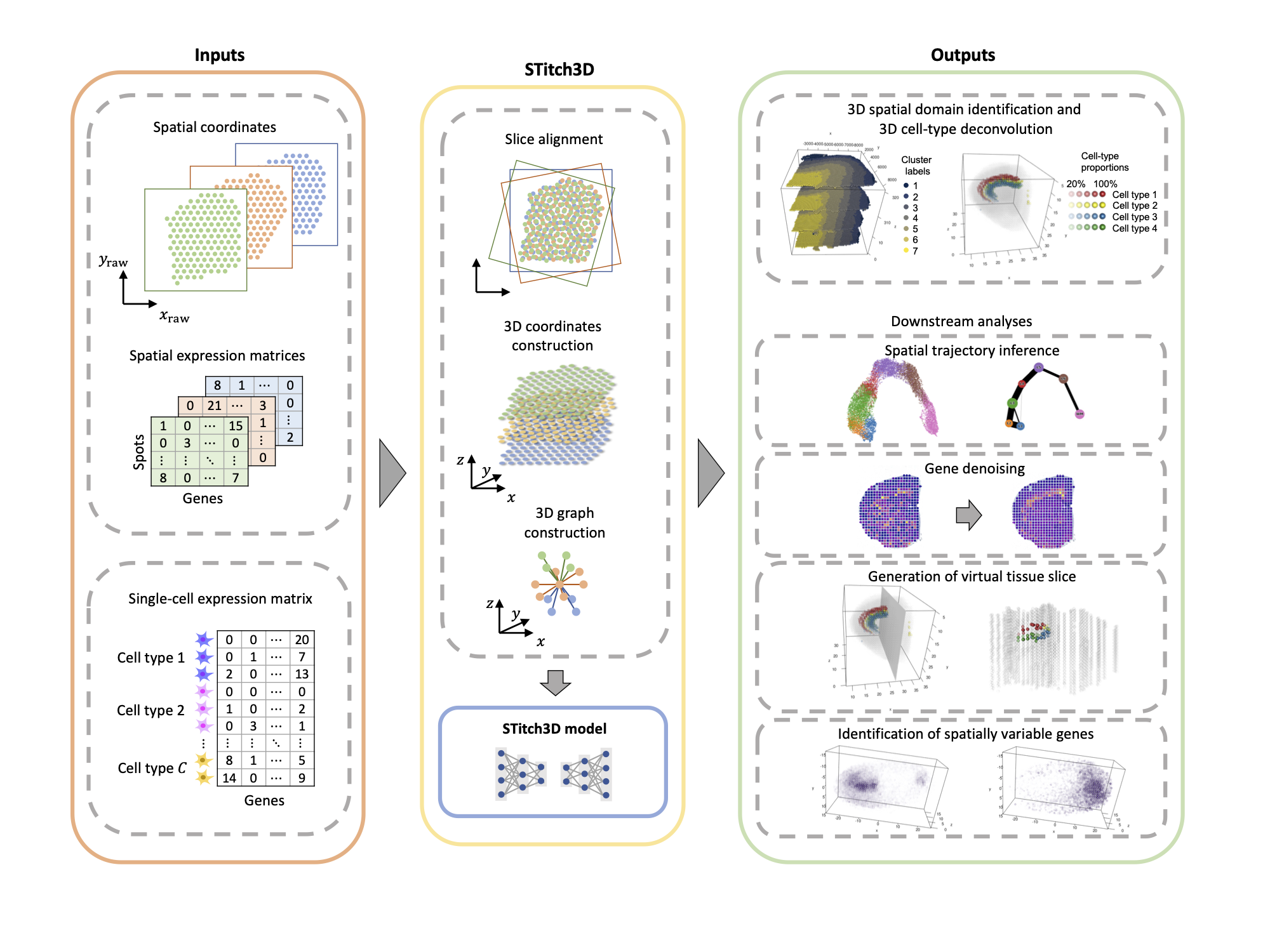
Construction of a 3D whole organism spatial atlas by joint modelling of multiple slices with deep neural networks Gefei Wang*, Jia Zhao*, Yan Yan, Yang Wang, Angela Ruohao Wu#, Can Yang#.
Nature Machine Intelligence. 2023.
Paper |
bioRxiv |
Software |
Website 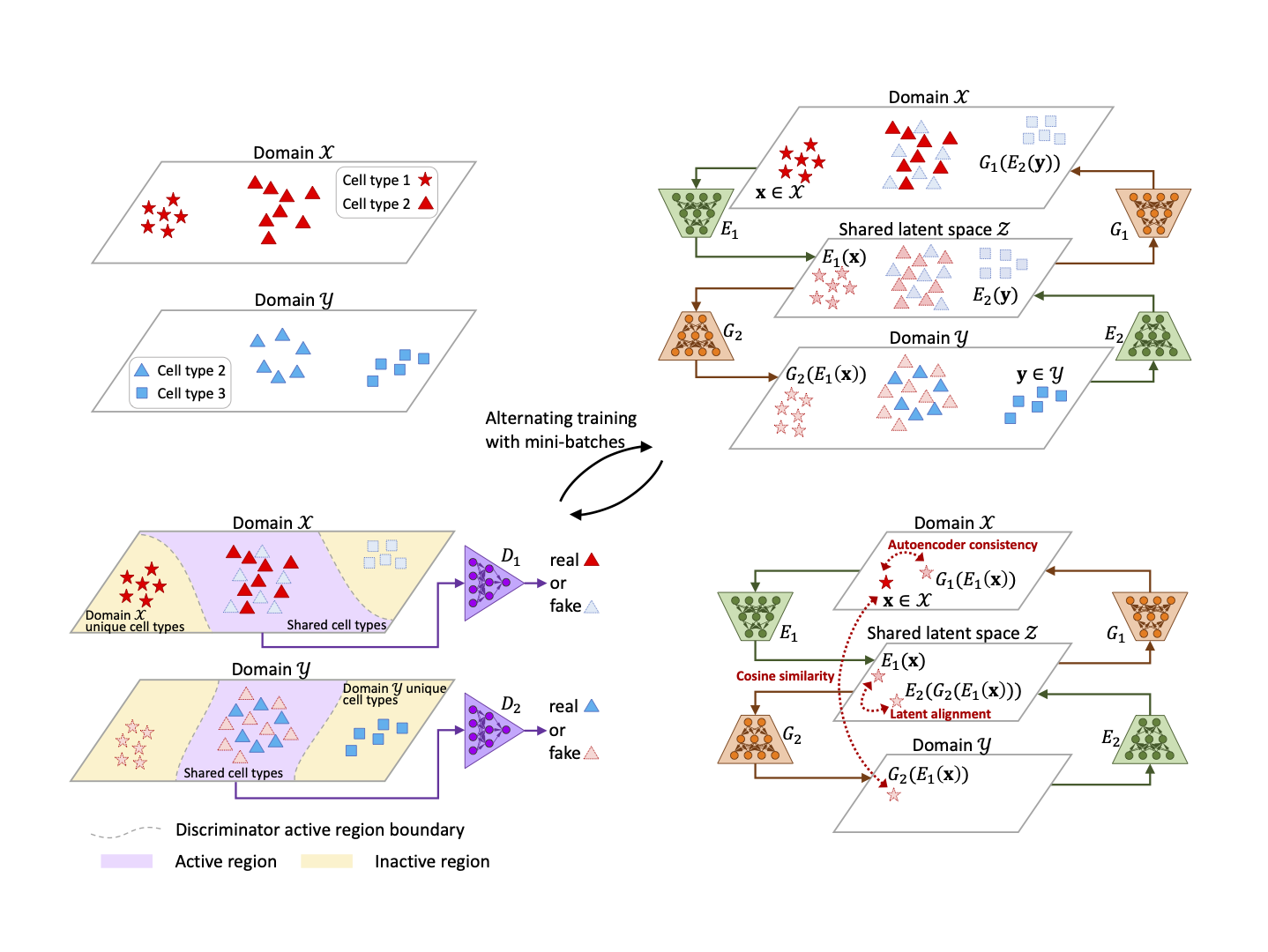
Adversarial domain translation networks for integrating large-scale atlas-level single-cell datasets Jia Zhao*,
Gefei Wang*, Jingsi Ming, Zhixiang Lin, Yang Wang,
The Tabula Microcebus Consortium†, Angela Ruohao Wu#, Can Yang#.
Nature Computational Science. 2022.
Paper |
bioRxiv |
Software |
Blog 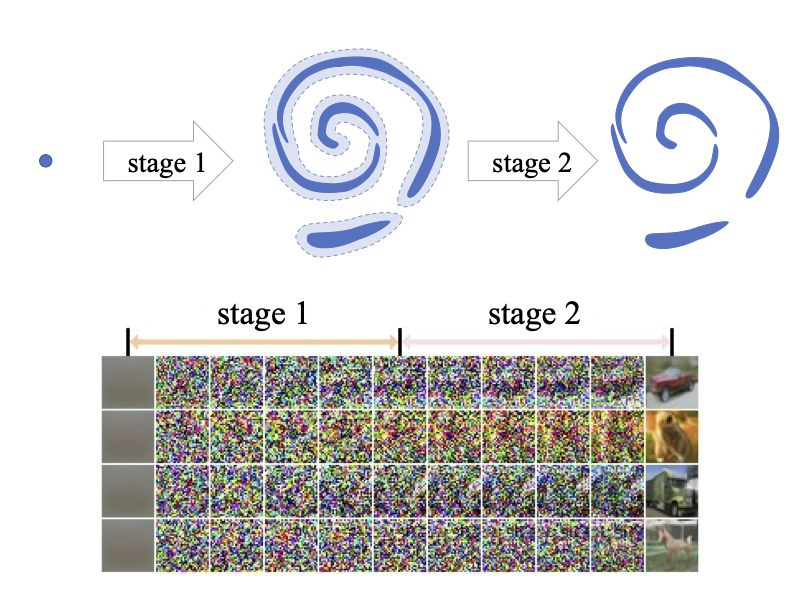
Deep generative learning via Schrödinger bridge Gefei Wang*, Yuling Jiao#, Qian Xu, Yang Wang, Can Yang#.
International Conference on Machine Learning. 2021.
Paper |
Code - Email: gefei.wang@yale.edu
- Address: 300 George Street, New Haven, CT 06511




